Explore the model and tools
• Forge: Experiment with ESM Cambrian using our hosted API.
• GitHub: Access ESM Cambrian code, model weights, and example workflows.
• SageMaker: Deploy and scale ESM Cambrian through AWS SageMaker.
AI is beginning to unlock an era of digital biology where we can use computation to go beyond the limits of experiments in the physical world. The universe of protein function is so vast and complex that we lack the ability to fully characterize it experimentally, and for many applications, we need to engineer functions that go beyond those in nature.
The physical limitations of performing experiments in the real world mean that unsupervised learning—algorithms that can learn from patterns in unlabeled data—is critical to unlocking the promise of digital biology. Discovering scalable techniques that can learn directly from protein sequences can illuminate biology at a level not yet possible with experimental characterization.
ESM Cambrian — Unsupervised learning for scientific discovery
ESM Cambrian (ESMC) is a parallel model family to our flagship ESM3 generative models. While ESM3 focuses on controllable generation of proteins for therapeutic and many other applications, ESMC focuses on creating representations of the underlying biology of proteins. ESMC scales up data and training compute to deliver dramatic performance improvements over ESM2.
ESM2 and its predecessor models have been used in a wide range of applications across basic scientific discovery, development of medicines, and sustainability, including a COVID-19 early warning system, state-of-the-art models for protein design, and antibody optimization. ESMC is a drop-in replacement for previous models that provides major improvements in both capability and efficiency.
ESMC models are available immediately for academic and commercial use under a new license structure designed to promote openness and enable scientists and builders.
ESMC is trained at three scales: 300M, 600M, and 6B parameters. We’re releasing ESMC 300M, and 600M as open weight models. ESMC 6B is available on Forge for academic use, and AWS Sagemaker for commercial use. ESMC will also be available on NVIDIA BioNemo soon. ESM models are always available for academic use for basic research.
Our frontier ESM3 models remain available for commercial use through partnerships to advance AI for drug discovery and other specific applications.
A preprint describing the new models is coming soon. In the meantime, get started building with ESMC. We’re excited to see what you create!
Defining a new frontier of performance
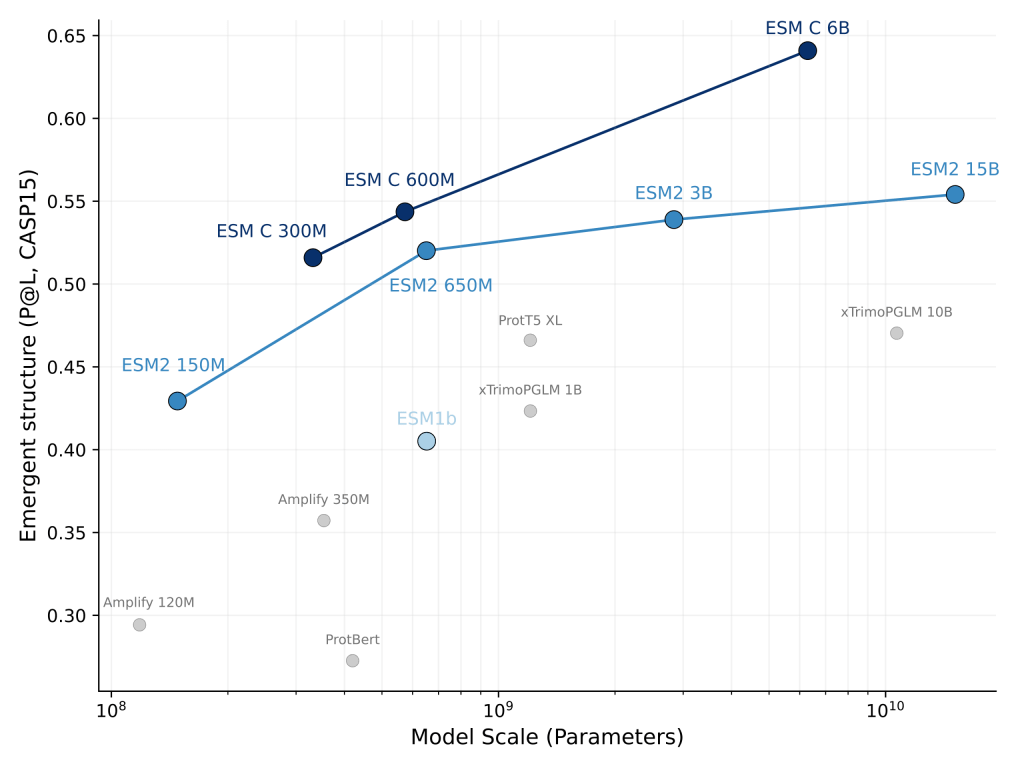
Since the introduction of ESM1, the first large transformer language model for proteins, ESM models have consistently defined the state of the art. Now we’re introducing a new generation of ESM models, ESMC, with major performance benefits over ESM2. ESMC achieves linear scaling across orders of magnitude in parameters, with each model matching, or even greatly exceeding, the performance of larger models of the previous generation. For example, the 300M parameter ESMC delivers similar performance to ESM2 650M with reduced memory requirements and faster inference. The 600M parameter ESMC rivals the 3B parameter ESM2 and approaches the performance of the 15B model. The 6B parameter ESMC sets a new benchmark, outperforming the best ESM2 models by a wide margin.
Learning biology from protein sequences
Language models are trained to predict the words or characters in a sequence. ESMC is trained using a masked language modeling objective, which requires the model to predict characters that have been masked out from their surrounding context.
Just as a person can fill in the blanks, such as:
To __ or not to __ that is the ___
We can train language models to fill in the blanks. Except in biology, instead of training the model to predict words, we train it to predict the characters in a protein sequence, i.e. its sequence of amino acids:
MSK_EEL_TG_VPILVELD_DVNGH_FSVS__GEGDA_YG
The patterns in protein sequences are linked to their underlying biological properties. Like a beam of light that shows an image of an object through its shadow, evolution encodes the biology of proteins into their sequences. This is because evolution is only free to select mutations to the sequence that are consistent with the biological function of the protein.
As language models learn to predict the patterns across many diverse protein sequences, information emerges within their internal representations that reflects the underlying biology of proteins. The information emerges through unsupervised learning, without the model ever seeing structure or function directly. Through the task of predicting amino acids, language models develop representations that reflect the hidden biological variables that determine which amino acids evolution chooses. The ability to learn structure and function from unlabeled sequence data means that language models can in principle scale far beyond existing AI approaches that only learn from structural or functional data.
Scaling laws
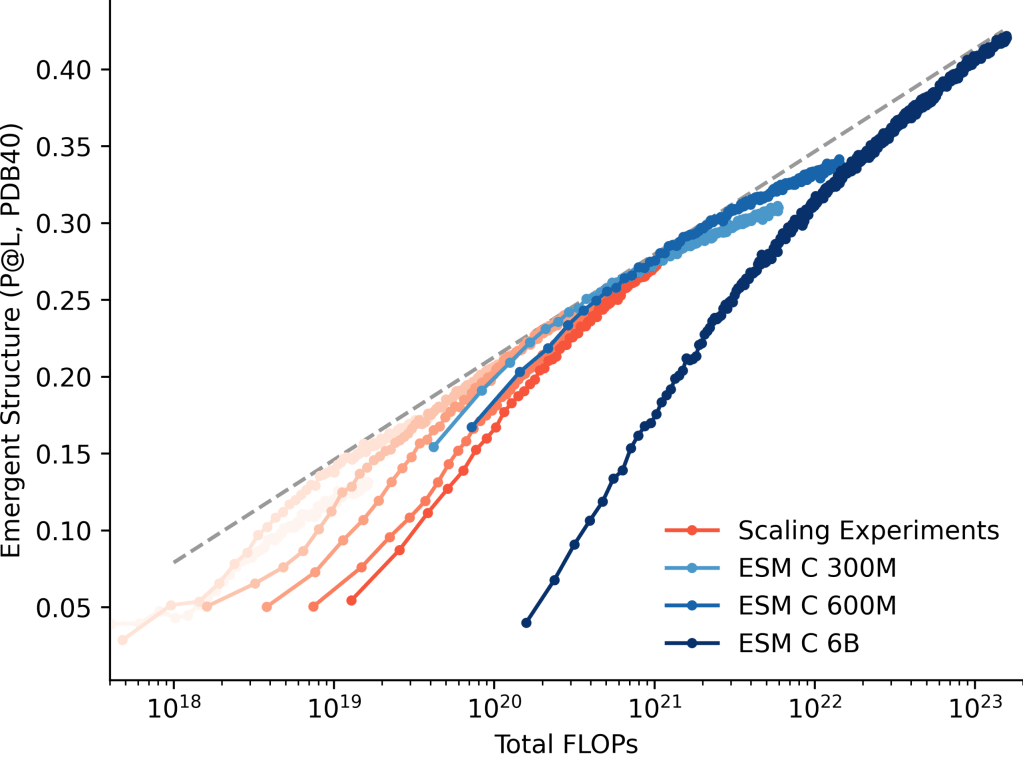
Scaling laws link the emergence of information about the biology of proteins with the scale of parameters, compute, and data. This allows the capabilities of models to be predicted as they are scaled up. ESMC demonstrates non-diminishing returns to scale up to 6 billion parameters, suggesting that continued scaling will result in further improvements.
Conclusion
ESMC is a step toward a more complete understanding of the deep and fundamental mysteries of life at the molecular level. As a practical technology, proteins have the potential to solve some of the most important problems facing our society from human health, to sustainability. Unlocking the secrets of protein molecules is making it possible to design proteins from first principles. ESMC is a foundational model for protein biology, one that can be used by builders across a wide range of applications, to imbue AI models with a deeper understanding of the biology of life’s most important and mysterious molecules.
Responsible development
We are committed to the responsible development of artificial intelligence for biology. ESMC was reviewed by a committee of scientific experts who concluded that the benefits of releasing the models greatly outweigh any potential risks. As we continue to push the frontier of AI for biology, we will continue to engage with the scientific community and stakeholders in government, civil society, and policy to ensure that we can maximize the benefits and minimize the potential risks to deliver the promise of this rapidly advancing new technology.
Training and evaluations
Training data
ESMC is trained on protein sequences from UniRef, MGnify, and the Joint Genome Institute (JGI). Sequence data is clustered at the 70% sequence identity level, resulting in 83M, 372M, and 2B clusters for UniRef, MGnify, and JGI, respectively.
Architecture
ESMC is based on the transformer architecture. It features Pre-LN, rotary embeddings, and SwiGLU activations. No biases are used in linear layers or layer norms.
| Params | 300M | 600M | 6B |
|---|---|---|---|
| # layers | 30 | 36 | 80 |
| width | 960 | 1152 | 2560 |
| # heads | 15 | 18 | 40 |
| Context length | 2048 | 2048 | 2048 |
| Learning rate | 5e-4 | 4.6e-4 | 3.1e-4 |
| Batch size in tokens | 4.2M | 4.2M | 4.2M |
| Num steps | 1.5M | 1.5M | 1.5M |
| Total tokens | 6.2T | 6.2T | 6.2T |
| FLOPs | 1.26e22 | 1.26e22 | 1.26e22 |
Training stages
ESMC is trained in two stages:
- Stage 1: For the first 1 million steps, the model uses a context length of 512, with metagenomic data constituting 64% of the training dataset.
- Stage 2: In the final 500,000 steps, the context length is increased to 2048, and the proportion of metagenomic data is reduced to 37.5%.
Evaluations
We use the methodology of Rao et al. to measure unsupervised learning of tertiary structure in the form of contact maps. A logistic regression is used to identify contacts. For a protein of length L, we evaluate the precision of the top L contacts (P@L) with sequence separation of 6 or more residues. The average P@L is computed on a temporally held-out set of protein structures (Cutoff date: May 1, 2023) for the scaling laws, and on CASP15 for the performance benchmark.
Citing ESMC
ESMC will be fully described in an upcoming preprint. This blog post will be updated with citation information as soon as the preprint is available. In the meantime, if necessary, you can cite:
“ESM Cambrian: Revealing the mysteries of proteins with unsupervised learning.” December 4, 2024. https://biohub.org/blog/esm-cambrian-unsupervised-learning